Publications
Find a full list of our publications at PubMed or Google Scholar; below are unpublished preprints and selected publications. Maurano Lab members are listed in bold.Preprints
Synthetic genomic dissection of enhancer context sensitivity and synergy
Ordoñez R, Ribeiro-dos-Santos AM, McLoughlin C, Ellis G, Ashe HJ, Zufiaurre NB, Brosh R, Majewski M, Camellato B, Boeke JD, Maurano MT*. bioRxiv (2025). doi: 10.1101/2025.08.13.669251Selected Publications
Iterative improvement of deep learning models using synthetic regulatory genomics.
Ribeiro-dos-Santos AM, Maurano MT*. Genome Research (2025). doi: 10.1101/gr.280540.125
Structure of a polymorphic repeat at the CACNA1C schizophrenia locus.
Moya R, Wang X, Tsien RW*, Maurano MT*. PNAS (2025). doi: 10.1101/2024.03.05.303780
Genomic context sensitizes regulatory elements to genetic disruption.
Ordoñez R, Zhang W, Ellis G, Zhu Y, Ashe HJ, Ribeiro-dos-Santos AM, Brosh R, Huang E, Hogan MS, Boeke JD, Maurano MT*. Molecular Cell (2024). doi: 10.1101/2023.07.02.547201
Genomic Analysis of a Synthetic Reversed Sequence Reveals Default Chromatin States in Yeast and Mammalian Cells.
Camellato B, Brosh R, Ashe HJ, Maurano MT, and Boeke JD. Nature 628:373-380 (2024).
The genetic basis of tail-loss evolution in humans and apes.
Xia B, Zhang W, Zhao G, Zhang X, Bai J, Brosh R, Wudzinska A, Huang E, Ashe H, Ellis G, Pour M, Zhao Y, Coelho C, Zhu Y, Miller A, Dasen JS, Maurano MT, Kim SY, Boeke JD, Yanai I. Nature 626: 1042–1048 (2024).
Mouse genome rewriting and tailoring of three important disease loci.
Zhang W, Golynker I, Brosh R, Fajardo A, Zhu Y, Wudzinska AM, Ordoñez R, Ribeiro-dos-Santos AM, Carrau L, Damani-Yokota P, Yeung S, Khairallah C, Gardner AV, Chalhoub N, Huang E, Ashe H, Khanna KM, Maurano MT, Kim SY, tenOever BR, and Boeke JD. Nature 623: 423–431 (2023).
Mouse genome rewriting with human DNA for disease modelling
Nature Reviews Genetic
Rewriting the mammalian genome, one locus at a time
Nature Genetics
Nature Reviews Genetic
Rewriting the mammalian genome, one locus at a time
Nature Genetics
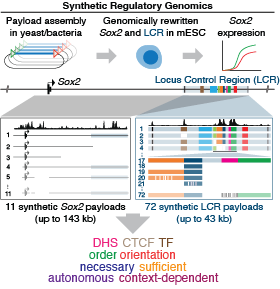
Synthetic regulatory genomics uncovers enhancer context dependence at the Sox2 locus.
Brosh R, Coelho C, Ribeiro-dos-Santos AM, Ellis G, Hogan MS, Ashe HJ, Somogyi N, Ordoñez R, Luther R, Huang E, Boeke JD, Maurano MT*. Molecular Cell 83: 1140-1152.e7 (2023).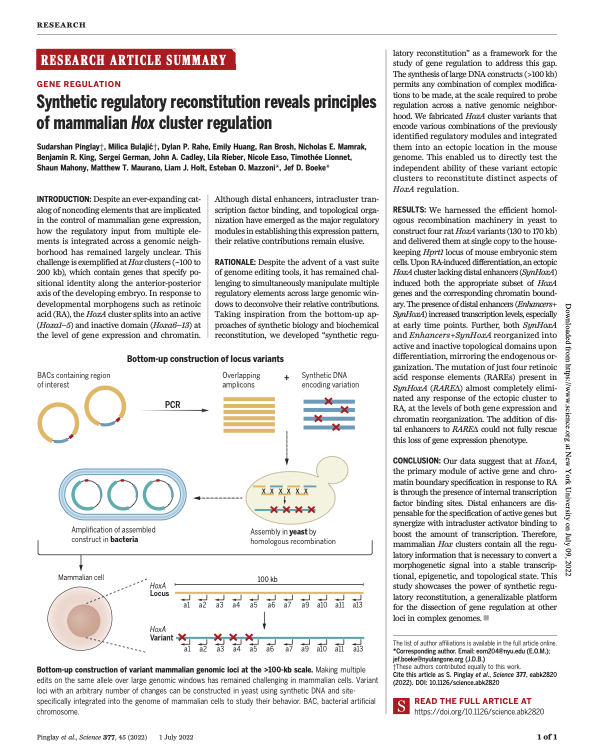
Synthetic genomic reconstitution reveals principles of mammalian Hox cluster regulation
Pinglay S, Bulajić M, Rahe DP, Huang E, Brosh R, German S, Cadley JA, Rieber L, Easo N, Mahony S, Maurano MT, Holt LJ, Mazzoni EO, Boeke JD. Science, 377: eabk2820 (2022).
Genomic context sensitivity of insulator function
Ribeiro-dos-Santos AM, Hogan MS, Luther RD, Brosh R, Maurano MT*. Genome Research. doi: 10.1101/gr.276449.121 (2022).
An Effector Index to Predict Causal Genes at GWAS Loci
Forgetta V, Jiang L, Vulpescu NA, Hogan MS, Chen S, Morris JA, Grinek S, Benner C, McCarthy MI, Fauman E, Greenwood CM*, Maurano MT*, and Richards JB*. Human Genetics. doi: 10.1101/2020.06.28.171561 (2022).
De novo mutations in childhood cases of sudden unexplained death that disrupt intracellular Ca2+ regulation
Halvorsen M, Gould L, Wang X, Grant G, Moya R, Rabin R, Ackerman MJ, Tester DJ, Lin PT, Pappas JG, Maurano MT, Goldstein DB, Tsien RW, Devinsky O. PNAS 118:e2115140118 (2021).
Geneticists find clues to sudden unexplained child deaths
Science
Genetics of sudden unexplained death in children
NIH Research Matters
Gene Mutations May Drive Sudden Unexplained Deaths in Children
Medscape
Science
Genetics of sudden unexplained death in children
NIH Research Matters
Gene Mutations May Drive Sudden Unexplained Deaths in Children
Medscape

Tissue context determines the penetrance of regulatory DNA variation
Halow J, Byron R, Hogan MS, Ordonez R, Groudine M, Bender MA, Stamatoyannopoulos JA*, and Maurano MT*. Nat. Communications. doi: 10.1038/s41467-021-23139-3 (2021).
A versatile platform for locus-scale genome rewriting and verification
Brosh R, Laurent JM, Ordoñez R, Huang E, Hogan MS, Hitchcock AM, Mitchell LA, Pinglay S, Cadley JA, Luther RD, Truong DM, Boeke JD, Maurano MT. PNAS. 118:e2023952118 (2021).
See Big-IN reagants on Addgene

Sequencing Identifies Multiple Early Introductions of SARS-CoV-2 to the New York City Region
Maurano MT*, Ramaswami S, Westby G, Zappile P, Dimartino D, Shen G, Feng X, Ribeiro-Dos-Santos AM, Vulpescu NA, Black M, Hogan M, Marier C, Meyn P, Zhang Y, Cadley J, Ordonez R, Luther R, Huang E, Guzman E, Serrano A, Belovarac B, Gindin T, Lytle A, Pinnell J, Vougiouklakis T, Boytard L, Chen J, Lin LH, Rapkiewicz A, Raabe V, Samanovic-Golden MI, Jour G, Osman I, Aguero-Rosenfeld M, Mulligan MJ, Cotzia P, Snuderl M*, and Heguy A*. Genome Research. doi: 10.1101/gr.266676.120 (2020)
Large-scale identification of functional variants impacting human transcription factor occupancy in vivo
Maurano MT*, Haugen E, Sandstrom R, Vierstra J, Kaul R, and Stamatoyannopoulos JA*. Nature Genetics 47: 1393–1401 (2015).
A new foundation for non-coding variant analysis
Nature Reviews Genetics Research Highlight
Additional supplemental files
Nature Reviews Genetics Research Highlight
Additional supplemental files
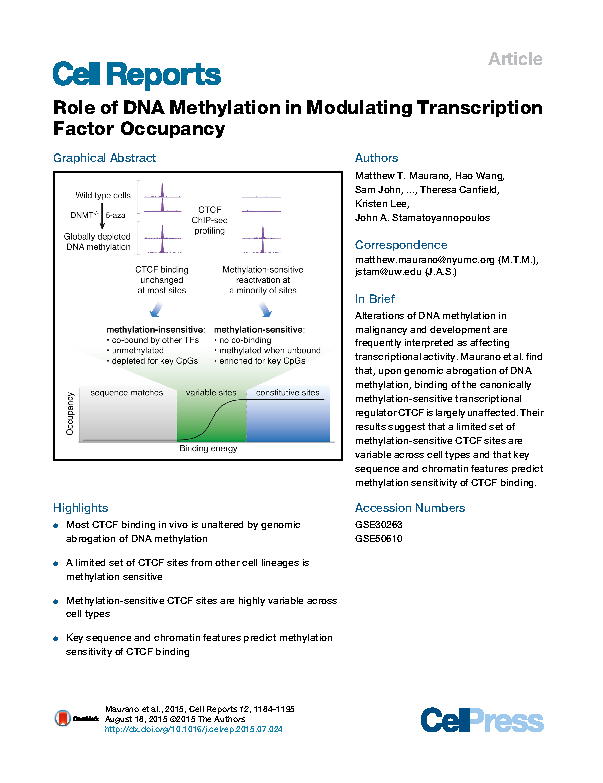
Role of DNA Methylation in Modulating Transcription Factor Occupancy
Maurano MT*, Wang H, John S, Shafer A, Canfield T, Lee K, and Stamatoyannopoulos JA*. Cell Reports 12: 1184–1195 (2015).
Genomic discovery of highly potent chromatin insulators for human gene therapy
Liu M, Maurano MT, Wang H, Song CZ, Navas PA, Emery DW, Stamatoyannopoulos JA and Stamatoyannopoulos G. Nature Biotech. 33: 198-203 (2015).
Insulating from genotoxicity
Nature Reviews Genetics Research Highlight
Nature Reviews Genetics Research Highlight

Systematic Localization of Common Disease-Associated Variation in Regulatory DNA
Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, Shafer A, Neri F, Lee K, Kutyavin T, Stehling-Sun S, Johnson AK, Canfield TK, Giste E, Diegel M, Bates D, Hansen RS, Neph S, Sabo PJ, Heimfeld S, Raubitschek A, Ziegler S, Cotsapas C, Sotoodehnia N, Glass I, Sunyaev SR, Kaul R, and Stamatoyannopoulos JA. Science 337: 1190–1195 (2012)
A GPS for Navigating DNA
Science Perspective
Regulatory regions
Nature Genetics Research Highlight
Additional supplemental files
Science Perspective
Regulatory regions
Nature Genetics Research Highlight
Additional supplemental files

Widespread Plasticity in CTCF Occupancy Linked to DNA Methylation
Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F, Lee K, Canfield T, Weaver M, Sandstrom R, Thurman RE, Kaul R, Myers RM, and Stamatoyannopoulos JA. Genome Research 22: 1680–1688 (2012).
The Accessible Chromatin Landscape of the Human Genome
Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, Giste E, Johnson AK, Johnson EM, Kutyavin T, Lajoie B, Lee B-K, Lee K, London D, Lotakis D, Neph S, Neri F, Nguyen ED, Qu H, Reynolds AP, Roach V, Safi A, Sanchez ME, Sanyal A, Shafer A, Simon JM, Song L, Vong S, Weaver M, Yan Y, Zhang Z, Zhang Z, Lenhard B, Tewari M, Dorschner MO, Hansen RS, Navas PA, Stamatoyannopoulos G, Iyer VR, Lieb JD, Sunyaev SR, Akey JM, Sabo PJ, Kaul R, Furey TS, Dekker J, Crawford GE, and Stamatoyannopoulos JA. Nature 489: 75–82 (2012).
An Expansive Human Regulatory Lexicon Encoded in Transcription Factor Footprints
Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, Maurano MT, Humbert R, Rynes E, Wang H, Vong S, Lee K, Bates D, Diegel M, Roach V, Dunn D, Neri J, Schafer A, Hansen RS, Kutyavin T, Giste E, Weaver M, Canfield T, Sabo P, Zhang M, Balasundaram G, Byron R, MacCoss MJ, Akey JM, Bender MA, Groudine M, Kaul R, and Stamatoyannopoulos JA. Nature 489: 83–90 (2012).
Widespread Site-Dependent Buffering of Human Regulatory Polymorphism
Maurano MT, Wang H, Kutyavin T, and Stamatoyannopoulos JA. PLoS Genetics 8: e1002599 (2012)
Interpretation of regulatory polymorphism
Nature Genetics Research Highlight
Complexities of occupancy and sequence
Nature Reviews Genetics Research Highlight
Nature Genetics Research Highlight
Complexities of occupancy and sequence
Nature Reviews Genetics Research Highlight